Global Journal of Medical and Clinical Case Reports
Training the Next Generation: A Generative AI-enhanced Framework for Teaching Bioinformatics to Life Science Students with Relevance to Cardiovascular Innovation
Camellia Moses Okpodu1* and Samelia Okpodu-pyuzza2
1Department of Botany, University of Wyoming, Laramie, WY 82070, USA
2The Giled Alliance Group, Huntingtown, MD 20639, USA
Cite this as
Okpodu CM, Okpodu-pyuzza S. Training the Next Generation: A Generative AI-enhanced Framework for Teaching Bioinformatics to Life Science Students with Relevance to Cardiovascular Innovation. Glob J Medical Clin Case Rep. 2025:12(6):140-142. Available from: 10.17352/2455-5282.000214Copyright License
© 2025 Okpodu CM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Bioinformatics is increasingly essential for life science students preparing for careers in biomedical research, biotechnology, and related fields. However, many undergraduate programs lack accessible entry points for teaching computational biology. This short communication presents a faculty-led instructional framework that integrates Python and generative AI tools to introduce bioinformatics in a life sciences curriculum. Designed for instructors without formal programming backgrounds, this approach emphasizes hands-on learning and interdisciplinary relevance. Our experience shows that generative AI can help reduce technical barriers for life science faculty and support scalable instruction in computational methods and data-centered biological exploration.
Introduction
As biological research becomes increasingly data-driven, life science undergraduates must develop foundational skills in bioinformatics [1]. Yet, many biology instructors lack the computational background to confidently teach these skills. This gap limits students’ exposure to essential tools used in genomics, systems biology, and biomedical research. Instructors in the biological sciences are increasingly expected to incorporate computational tools into their teaching, yet many feel unprepared to do so [2]. The rapid expansion of computational biology, coupled with the accelerating pace of technological change, can make the field appear daunting, especially to educators without formal training in computer science [3]. This perceived barrier often leads to missed opportunities for students to engage with essential skills in data analysis, modeling, and scientific computing [4].
As cardiovascular medicine increasingly relies on genomic and multi-omics data to drive precision diagnostics and therapeutics, there is a growing need for a workforce trained in bioinformatics and data science. Life science undergraduates, many of whom will pursue careers in biomedical research or clinical support roles, must be equipped with foundational computational skills to contribute meaningfully to this evolving landscape.
To address this challenge, we developed a pilot instructional module that integrates generative Artificial Intelligence (AI) with open-source bioinformatics platforms. Our goal was to develop a practical and scalable instructional model that empowers biology educators, especially those without formal training in computer science, to confidently integrate computational biology into their courses. By leveraging accessible tools and generative AI, this approach supports faculty in delivering interdisciplinary content that aligns with emerging trends in biomedical and life science research.
Methods
To ensure relevance to biomedical and clinical contexts, we selected datasets and case examples related to human physiology. Our module development provided a bridge between computational exercises and real-world applications in cardiovascular research.
Course context
To ensure relevance to biomedical and clinical contexts, we developed an instructional module focused on human physiology, using real-world blood pressure datasets. This module introduced students to basic data analysis techniques while exploring cardiovascular function. By integrating computational exercises with physiological data, we created a bridge between foundational biology instruction and contemporary biomedical applications which demonstrates how instructors can incorporate bioinformatics into life science courses without requiring advanced programming expertise. Oxygen saturation (SpO2) is a highly relevant and accessible physiological measure for cardiovascular studies, especially in the context of exercise physiology, hypoxia, and circulatory efficiency. It’s also non-invasive and commonly used in both clinical and research settings. The module was created for an undergraduate course for life science students a public university.
Tools and content
We utilized the open-source Python, which provided a user-friendly platform for managing dependencies and executing code. Data analysis was performed using Python libraries such as NumPy and Pandas, which enabled efficient handling and processing of physiological data. The dataset, downloaded in.csv format from the PhysioNet database, included synchronized measurements of SpO2, heart rate, and blood pressure, allowing for correlation analysis within a cardiovascular context [5,6]. This data is also stored in GitHub.
AI Integration and Generating Code – We used Bing GPT-3 both help generate example code and troubleshoot errors. The code we used to visualize a heat map of our selected data can be found at https://github.com/cokpodu/AIPhys.
Instructor preparation
Faculty used AI tools to develop instructional materials, simulate coding workflows, and model problem-solving strategies during class sessions. The biggest problem we encountered is that we often introduced spacing in our code. We used Bing to help us identify where those spaces occurred.
Results
This module was developed for an introductory life science course focused on human physiology, with the specific intention of being delivered in an online format. To bridge the gap between biological content and computational instruction, we incorporated generative AI tools into the course design to assist life science instructors, particularly those without extensive training in computer science, in integrating basic programming and data analysis techniques. These tools enabled faculty to write and troubleshoot Python code with greater confidence, particularly by identifying syntax errors and supporting the construction of visualizations.
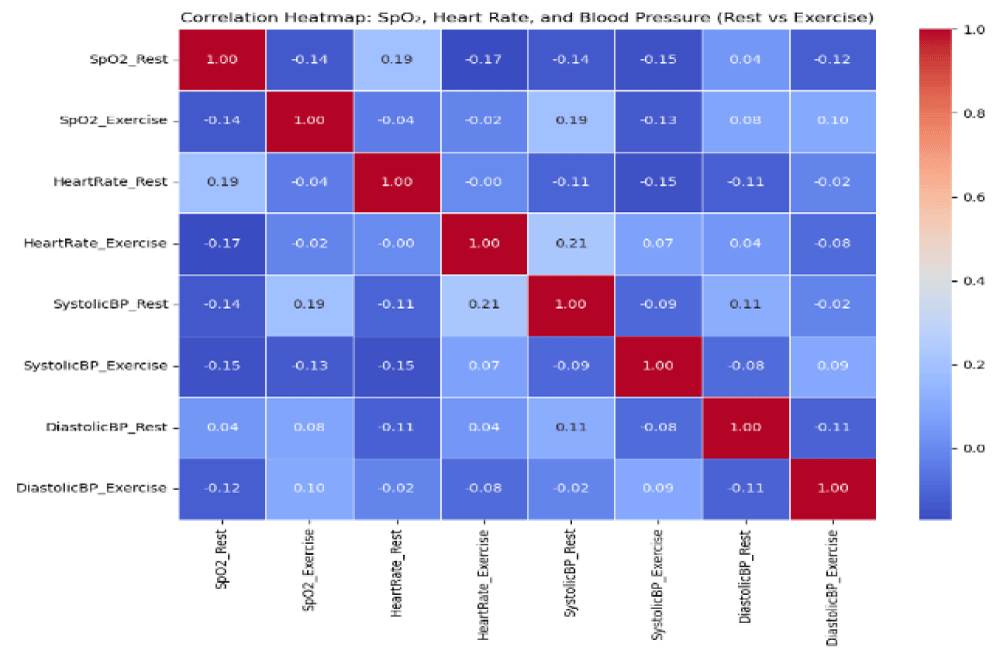
One key instructional output was a correlation matrix, generated using Python and shown in Figure 1. This module was created to empower instructors to move beyond basic coding and begin interpreting physiological relationships in their data. The visualization allowed educators to analyze patterns across variables such as SpO2, heart rate, and blood pressure. For example, a strong positive correlation between resting SpO2 and light exercise values (r = 0.89) reaffirmed known physiological responses that oxygen saturation remains relatively stable at low activity levels. As exercise intensity increased, correlations decreased (r = 0.74–0.78 for moderate, and r = 0.58–0.62 for high intensity), which aligns with expected variability in desaturation as physiological demand increases. A slight inverse correlation between resting SpO2 and heart rate during exercise (r =-0.17) further reflects common compensatory mechanisms of cardiovascular function under strain. Recovery values also showed strengthened correlations (e.g., r = 0.81 between moderate exercise and recovery), suggesting more efficient oxygen regulation in individuals who sustained better SpO2 during exertion.
These findings are in line with the physiological mechanisms discussed by Yunusa [7], who emphasized the dynamic relationship between oxygen saturation and individual cardiorespiratory efficiency across different exercise intensities. By using generative AI to both generate and interpret this output, instructors are able to connect computational workflows directly to meaningful biological insights, thereby building both coding fluency and pedagogical confidence for integrating bioinformatics into life science instruction.
Building on this approach, we have integrated the principles from the module into our upper-level Computational Biology course, designed for advanced undergraduates and beginning graduate students across disciplines. The course attracts students from majors such as botany, computer science, chemical engineering, mathematics, plant sciences, and zoology, many of whom enter with little to no programming experience. By anchoring instruction in real-world biological questions, the course emphasizes practical applications of bioinformatics, statistics, and computational tools. Each section introduces a biological problem and guides students through relevant analytical methods using real datasets. The course fulfills interdisciplinary elective requirements and requires only introductory biology and calculus or statistics as prerequisites. Its primary goals are to (1) demonstrate the essential role of computation in modern biology, (2) foster conceptual understanding of common analytical frameworks, and (3) build transferable skills in coding, data interpretation, and critical thinking.
Discussion
The development of this module shows that generative AI can serve as a valuable support tool for instructors who are introducing bioinformatics into life science education. By lowering technical barriers, AI made it easier for faculty without programming backgrounds to incorporate Python-based coding exercises and physiological data analysis into their teaching. In this cardiovascular-focused module, instructors used AI-assisted tools to generate a correlation matrix of physiological variables and interpret their biological meaning. Visualizations of SpO2 dynamics highlighted important trends, such as decreased correlations with increasing exercise intensity and stronger relationships during recovery. These findings supported discussions around oxygen transport, cardiovascular adaptation, and individual variation in desaturation and reoxygenation.
Recent advancements in cardiovascular research have demonstrated the power of integrating multi-omics data with AI-driven analysis to uncover novel biomarkers, predict disease progression, and personalize treatment strategies [8]. For example, machine learning models applied to genomic, transcriptomic, and proteomic data have shown promise in identifying molecular signatures of heart failure and atherosclerosis [8]. By introducing instructors and, ultimately, their students to the tools and thinking behind these innovations, our framework contributes to broader engagement with data-intensive cardiovascular science. This experience illustrates that generative AI is more than a tool for code generation. It can also help instructors bridge the gap between computational outputs and biological interpretation. By supporting both visualization and pattern recognition, AI helped faculty engage more deeply with physiological concepts while building confidence in their ability to teach bioinformatics in a meaningful and accessible way.
- Zawacki-Richter O, Marín VI, Bond M, Gouverneur F. Systematic review of research on artificial intelligence applications in higher education: Where are the educators? Int J Educ Technol High Educ. 2019;16(1):39. Available from: https://doi.org/10.1186/s41239-019-0171-0
- Alpaydin E, Aydin M. Biological computation and computational biology: Survey, challenges, and perspectives. Artif Intell Rev. 2020;53:1–28. Available from: https://doi.org/10.1007/s10462-020-09951-1
- Viet NK. The use of generative AI tools in higher education: Ethical and pedagogical principles. J Acad Ethics. 2025. Available from: https://doi.org/10.1007/s10805-025-09607-1
- Mehrgardt P, Khushi M, Poon S, Withana A. Pulse Transit Time PPG Dataset (version 1.1.0). PhysioNet. 2022. RRID:SCR_007345. Available from: https://doi.org/10.13026/jpan-6n92
- Goldberger A, Amaral L, Glass L, Hausdorff J, Ivanov PC, Mark R, et al. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation [Online]. 2000;101(23):e215–e220. Available from: https://doi.org/10.1161/01.cir.101.23.e215
- Lin M, Guo J, Gu Z, Tang W, Tao H, You S, et al. Machine learning and multi-omics integration: Advancing cardiovascular translational research and clinical practice. J Transl Med. 2025;23:Article 388. Available from: https://doi.org/10.1186/s12967-025-06425-2
- Yunusa HH. The impact of physical activity on oxygen saturation levels. Int J Physiol Health Phys Educ. 2023;5(1):21–3. Available from: https://doi.org/10.33545/26647265.2023.v5.i1a.53
- Khera R, Topol EJ. The potential of AI in cardiovascular care: Focus of review. J Am Coll Cardiol (JACC) – State-of-the-Art Review. 2024 Jun 24. Available from: https://www.acc.org/Latest-in-Cardiology/Articles/2024/06/24/16/51/The-Potential-of-AI-in-Cardiovascular-Care-Focus-of-Review